Advantages of functional analysis in comparison of different chemometric techniques for selecting obesity-related genes of adipose tissue from high-fat diet-fed mice
DOI:
https://doi.org/10.15419/bmrat.v8i3.666Keywords:
gene ontology, obesity, principal component analysis, protein-protein interaction, random forestAbstract
Introduction: Obesity is a lifestyle disease that is becoming prevalent nowadays and is associated with a surplus in energy balance related to lipid metabolism, inflammation and hypoxic condition, resulting in maladaptive adipose tissue expansion. This study used the publicly available gene dataset to identify a small subset of important genes for diagnostics or as potential targets for therapeutics.
Methods: Chemometric analyses by principal component analysis (PCA), random forest (RF), and genetic algorithm (GA) were used to identify 50 genes that differentiate adipose samples from high-fat diet- and normal diet-fed mice. The first 30 important genes were studied for classifying the samples using six different classification techniques. Gene ontology (GO), pathway analysis, and protein-protein interaction studies on the 50 selected genes were subsequently done to identify important functional genes. Finally, gene regulatory effects by microRNA were assessed to confirm the genes’ potential as targets for new therapeutic drugs.
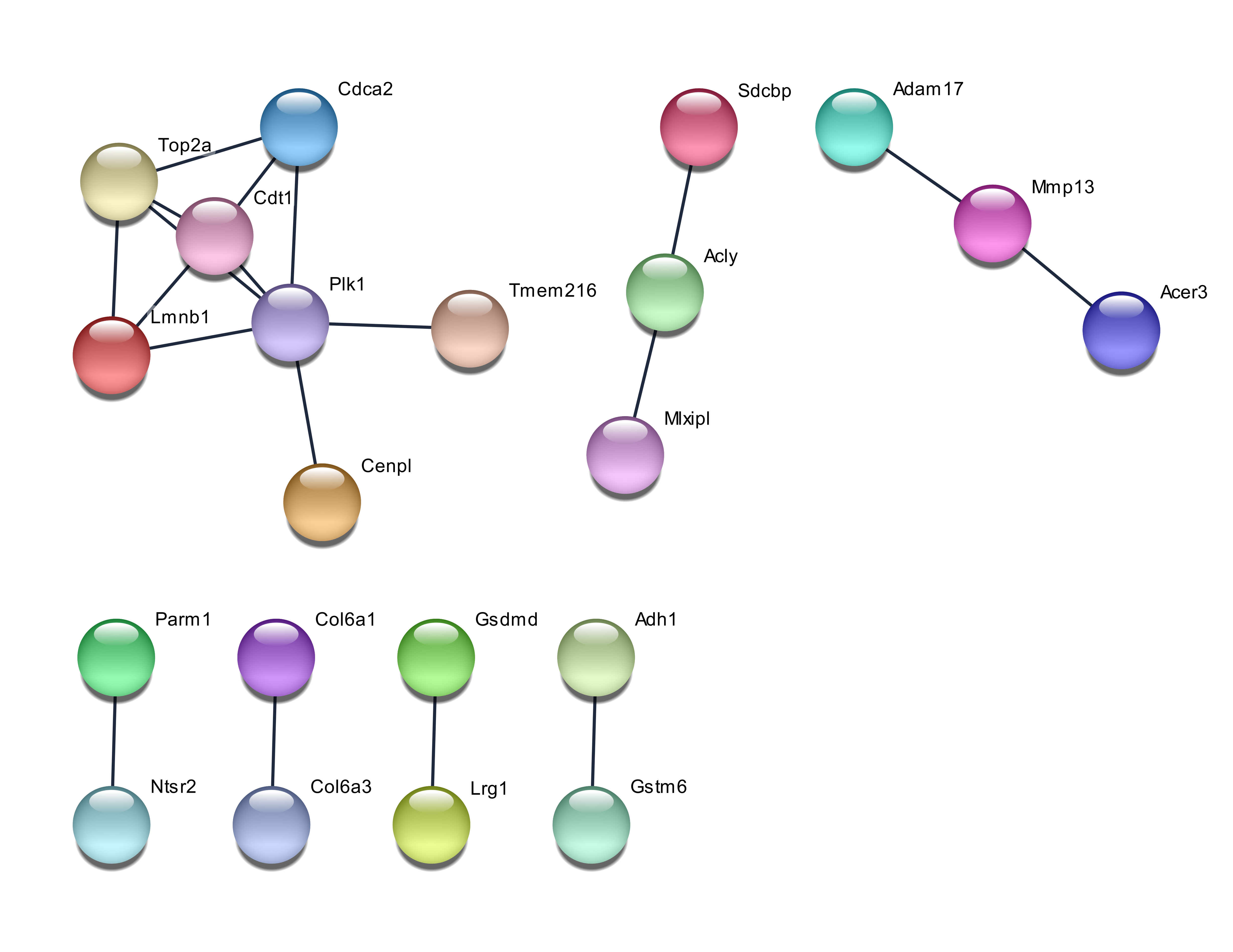
Results: The genes identified by RF are best for differentiating the samples, followed by PCA, with the least predictability shown by genes chosen by GA. However, PCA identified more genes with functional importance, such as the hub genes ATP5a1 and Apoa1. ATP5a1 is the main hub gene, whereas Apoa1 is involved in cholesterol metabolism. Vapa and Npc2 are crosstalk genes that link both of these main genes and could be targeted for therapeutic drug design.
Conclusion: The combination of different chemometric techniques and functional analysis of genes could be used to select for a small number of genes which could serve as more suitable diagnostic or therapeutic targets.
Downloads
Published
Issue
Section
License
Copyright The Author(s) 2017. This article is published with open access by BioMedPress. This article is distributed under the terms of the Creative Commons Attribution License (CC-BY 4.0) which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
