In silico and in vitro analysis of differentially expressed microRNAs, circular RNAs, and p53 in bortezomib-resistant multiple myeloma
DOI:
https://doi.org/10.15419/bmrat.v9i7.754Keywords:
Multiple myeloma, Bortezomib resistance, Bioinformatics analysis, miRNA, Circular RNA, p53Abstract
Background: Multiple myeloma (MM), which constitutes 1% of all cancers, remains incurable due to the acquisition of drug resistance. Proteasome inhibitor therapy such as bortezomib (BTZ), one of the key therapies used to treat MM, is impeded by the occurrence of BTZ resistance. The precise mechanism underlying this acquired BTZ resistance in MM is not yet well understood. In recent times, the role of non-coding RNAs, such as microRNAs (miRNAs) and circular RNAs (circRNAs), in cancer progression and drug resistance has been demonstrated. The present study aimed to identify differentially expressed miRNAs (DEmiRs), circRNAs, and target genes in BTZ-resistant MM in silico and in vitro.
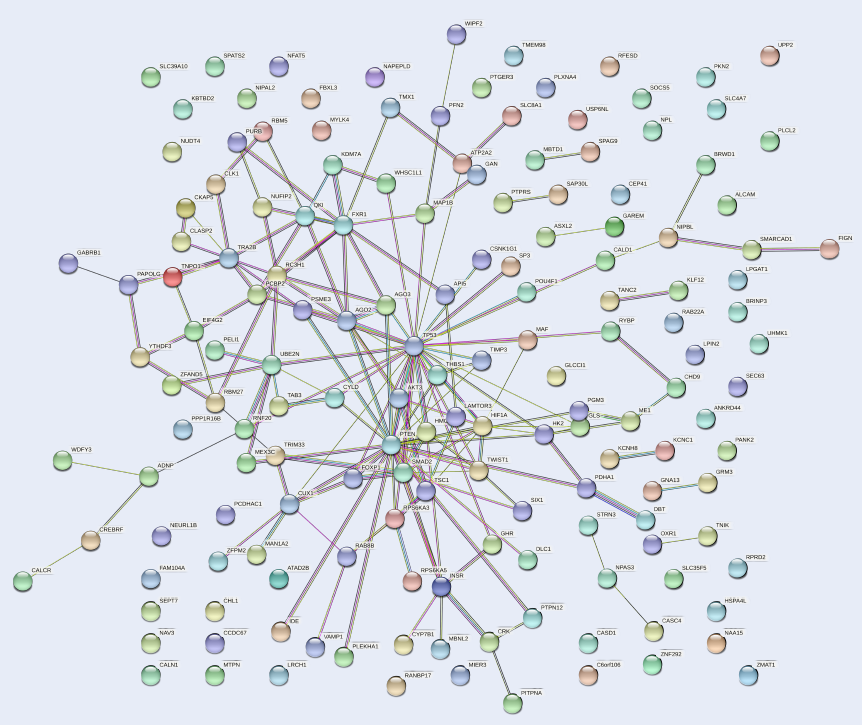
Methods: DEmiRs were obtained and analyzed from microarray datasets of MM patients from the Gene Expression Omnibus (GEO) database using GEO2R. Targets of a DEmiR were detected using mirDIP software. Gene ontology and pathway enrichment analysis of the targets were executed using the DAVID database. A protein–protein interaction network of targets was constructed and analyzed using the STRING database and Cytoscape tool. Expression levels of a DEmiR and an interacting circRNA were assessed by qRT-PCR. Target gene expression was assessed by qRT-PCR and western blot.
Results: Among the identified DEmiRs, hsa-mir-151a-3p was found to be upregulated in BTZ-resistant MM. One circRNA, hsa_circ_0000073, that was predicted to interact with hsa-mir-151a-3p using the CircInteractome platform, was downregulated in BTZ-resistant MM. Among the targets of hsa-mir-151a-3p, p53 was downregulated at both the transcript and protein levels in BTZ-resistant MM.
Conclusions: This study indicates that these molecular players could be implicated in BTZ-resistant MM.
Published
Issue
Section
License
Copyright The Author(s) 2017. This article is published with open access by BioMedPress. This article is distributed under the terms of the Creative Commons Attribution License (CC-BY 4.0) which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
