Evaluation of UHRF1 and P16INK4A expression levels in newly diagnosed AML patients
DOI:
https://doi.org/10.15419/bmrat.v5i9.475Keywords:
Acute myelogenous leukemia, Gene expression, p16INK4A, UHRF1Abstract
Introduction: Gene mutation is an infrequent cause of tumor suppressor gene (TSG) defect in de novo AML patients. Instead, it seems that leukemic cells employ epigenetic tricks to attenuate the negative impacts of intact TSGs. Ordinarily, critical TSGs, such as p16INK4A, is hyper-methylated in AML blasts under the impact of master epigenetic regulators, such as UHRF1. In this study, we investigated the correlation between UHRF1 and p16INK4A gene expression levels in newly diagnosed AML patients.
Methods: Bone marrow and peripheral blood samples were obtained from 50 newly diagnosed AML patients and 18 healthy normal control subjects. Gene expression levels of UHRF1 and P16INK4A were surveyed using SYBR Green Quantitative Real-time PCR. Statistical analyses were done using SPSS statistical software 21.0.
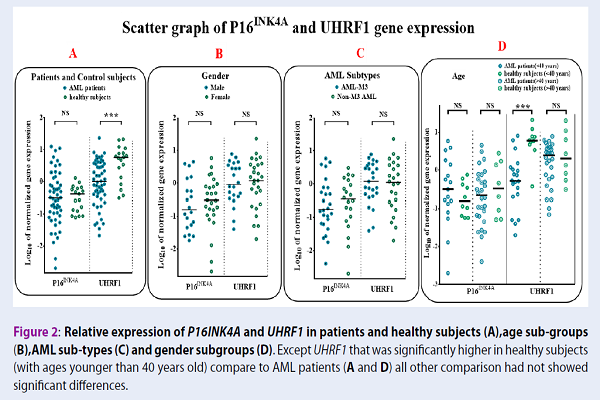
Results: P16INK4A gene expression showed reduced levels in 80.64% of patients above 45 years of age, while only 32% of patients below 45 years had reduced expression levels. The Spearman correlation test also demonstrated a significant negative correlation between UHRF1 and p16INK4A gene expression levels in AML patients, which was not observed in the control group (r=0.343 and P= 0.015).
Conclusion: Regarding the age-related patterns of UHRF1 and p16INK4A gene expression, and also the presence of negative correlation between them, we conclude that UHRF1 may potentially be involved in p16INK4A down-regulation in elderly AML patients, which may subsequently facilitate the progression of AML in older ages.
Downloads
Published
Issue
Section
License
Copyright The Author(s) 2017. This article is published with open access by BioMedPress. This article is distributed under the terms of the Creative Commons Attribution License (CC-BY 4.0) which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
