Genetic variations in the Orf7a protein of SARS-CoV-2 and its possible role in vaccine development
DOI:
https://doi.org/10.15419/bmrat.v8i8.688Keywords:
COVID-19, Epitope, Mutation, Orf7a, SARS-CoV-2, VaccineAbstract
Introduction: SARS-CoV-2 (Severe Acute Respiratory Syndrome) is the causative agent of the novel coronavirus disease (COVID-19) that has been creating an unprecedented situation globally. The recurrent mutations in SARS-CoV-2 genomes impact on the vaccine designing strategies. The Orf7a is a 121-amino acid-long type I transmembrane accessory protein encoded by the genome of SARSCoV- 2 and plays a crucial role in the virus–host interaction. The present study aimed to analyze the variations occurring in Orf7a due to multiple mutations and its immunological role in developing a promising therapeutic target to curb SARS-CoV-2 infections.
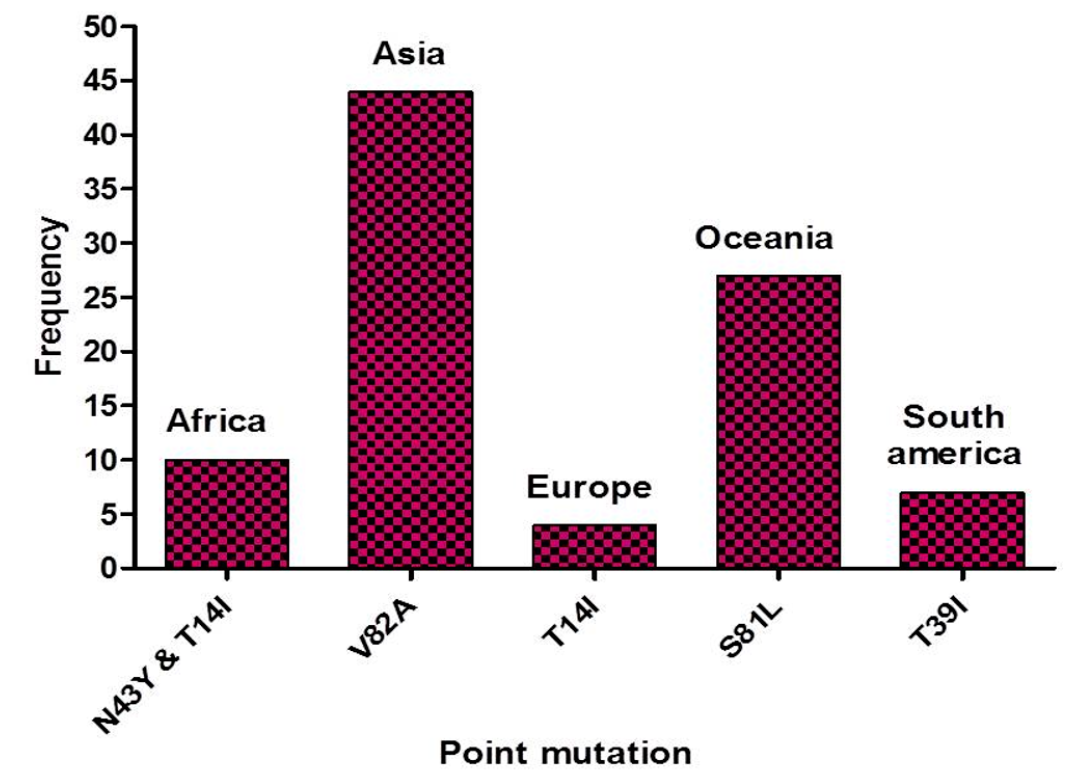
Methods: 16,161 sequences of Orf7a reported from the onset of this disease until 13 June 2021 from five continents were compared to identify genetic variations in the protein.
Results: A total of 470 point mutations were detected in the sequences submitted. Subsequently, the nature of mutations (deleterious or neutral) was determined. Furthermore, the physicochemical properties, antigenicity, allergenicity, toxicity, and stability of Orf7a protein were estimated to demonstrate the stability of the protein. Additionally, we identified three B-cell immune epitopes, and their MHC cluster analysis was also performed.
Conclusion: The recurrent mutations in Orf7a of SARS-CoV-2 provide a deep understanding of its role in the virus–host interactions. Findings of our study revealed that the predicted epitopes could be promising candidates for a vaccine against COVID-19 infections.
Downloads
Published
Issue
Section
License
Copyright The Author(s) 2017. This article is published with open access by BioMedPress. This article is distributed under the terms of the Creative Commons Attribution License (CC-BY 4.0) which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
