Motif prediction of abemaciclib in a breast cancer cell line using ChIP-Seq data analysis
DOI:
https://doi.org/10.15419/bmrat.v9i3.732Keywords:
ChIP-Seq, Motif, Binding site, Abemaciclib, Galaxy, MCF-7Abstract
Introduction: Chromatin immunoprecipitation sequencing (ChIP-Seq) is a DNA sequencing technique for the identification of binding sites in genomic sequences. ChIP-Seq experiments are a combination of immunoprecipitation and sequencing techniques that are used for the identification of chromatin regions that bind molecules such as transcription factors (TFs), histones, and drugs. In this study, computational analysis of ChIP-Seq data was performed to predict the binding sites in breast cancer cells and their association with several molecules, such as TFs and drugs. A complete and comprehensive computational study has been performed to predict the binding sites of abemaciclib. Functional enrichment of selected motifs was performed to identify important motifs that function in breast cancer and show binding with the drug abemaciclib.
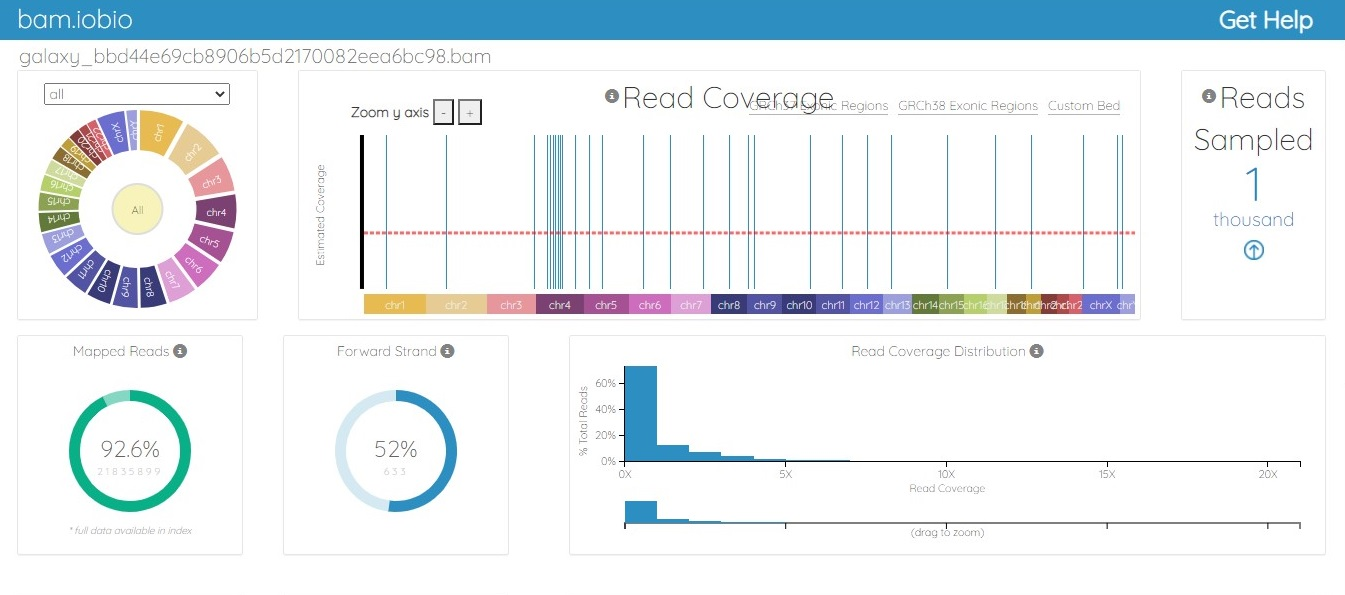
Methods and Materials: The ChIP-Seq analysis protocol was performed using the Galaxy server (https://usegalaxy.org/). The abemaciclib binding motif was identified using MEME tools. For this research, ChIP-Seq data from a breast cancer cell line was retrieved from the GEO database, accession number GSM4763932. This dataset includes ChIP-Seq of MCF-7 cells exposed to abemaciclib. The ENA browser was used to retrieve the data. Statistical analysis was performed using the default parameters of Fast-QC, Multi-QC, Map with BWA, Filter SAM and BAM, MACS2, and ChIP Seeker tools on the Galaxy server.
Results: Computational analysis identifies the abemaciclib consensus binding sequence as TGGCTCACGCCTGTAATCCCAGCACTTT, and this motif occurs 2980 times in the Homo sapiens reference genome hg19.
Conclusions: This study identifies the binding sites and affinity of abemaciclib in a breast cancer cell line.
Published
Issue
Section
License
Copyright The Author(s) 2017. This article is published with open access by BioMedPress. This article is distributed under the terms of the Creative Commons Attribution License (CC-BY 4.0) which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
