Optimizing RNA extraction and library preparation from oral squamous cell carcinoma FFPE samples for next-generation RNA sequencing
DOI:
https://doi.org/10.15419/bmrat.v10i10.840Keywords:
FFPE, formalin-fixed paraffin-embedded, next-generation sequencing, oral squamous cell carcinoma, RNA sequencingAbstract
Introduction: Formalin-fixed paraffin-embedded (FFPE) tissue provides a valuable source of information for pathological studies and oral cancer pathology. However, FFPE tissue immobilization and storage often cause the partial degradation of nucleic acids, resulting in mRNA sequencing libraries that may not be of sufficient quantity and quality for gene expression analysis. We optimized the RNA extraction and library preparation process to increase the amount of useful data obtained with low-quality RNA from FFPE oral cancer tissue samples.
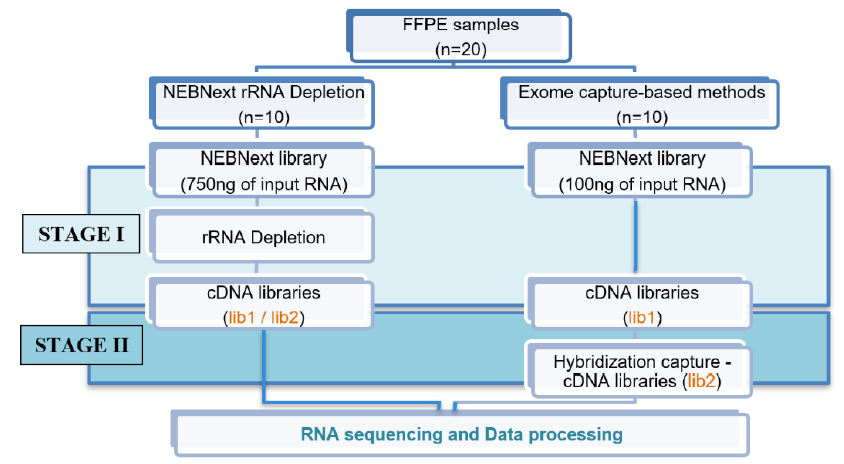
Methods: This study used 20 samples stored for 1 - 2 years. After RNA extraction from FFPE samples, we compared two methods for library preparation, rRNA depletion, and exome capture, to make recommendations for metrics such as RNA input and output concentrations and generated full RNA sequencing data for downstream bioinformatics analysis.
Results: The quantity of RNA extracted from six 8-mm-thick slices of FFPE tissue was sufficient for library preparation (130 ng/µL); sample quality did not differ significantly with storage time. Additionally, the RNA samples had an average DV200 index of 30% - 50%. Exome capture outperformed rRNA depletion for library preparation in library output concentration (p < 0.001) and RNA sequencing data generated for bioinformatics analysis.
Conclusion: RNA can be extracted from FFPE samples for sequencing, provided they have been handled and stored appropriately. Exome capture is the best method for preparing libraries for RNA sequencing from low-quality tissue samples such as FFPE.
Published
Issue
Section
License
Copyright The Author(s) 2017. This article is published with open access by BioMedPress. This article is distributed under the terms of the Creative Commons Attribution License (CC-BY 4.0) which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
